Last updated: February 04, 2015
Jan 6 2015 Next Phase Nhgris Genome Sequencing Program
Next Phase: NHGRI's Genome Sequencing Program
January 5, 2015
Large-scale genome sequencing has been a central component of NHGRI's Extramural Research Program since the Institute's inception, starting with the Human Genome Project. Considering the time and money it took to generate that first reference human genome sequence (upwards of 8 years and a billion dollars, respectively), I am amazed that we are now on the cusp of being able to generate a human genome sequence in a day and for less than $1,000. That is simply astonishing!
Riding the waves of the many technical advances in DNA sequencing, the well-established NHGRI Genome Sequencing Program (GSP) has tackled a variety of biomedical research projects. Over the years, its priorities and focus areas have continuously 'morphed' to capitalize on new opportunities- starting with sequencing that first human genome and then moving on to sequence the genomes of multiple organisms for comparative genomics studies, sequencing many human genomes for human genomic variation studies, sequencing many cancer genomes for cancer genomics studies, and, most recently, sequencing human genomes for studies of rare and common diseases.
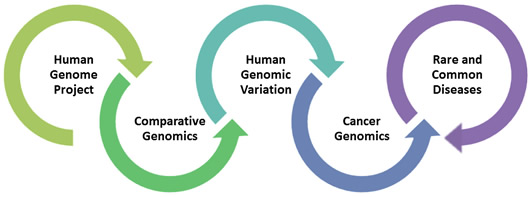
With the funding period of the current set of GSP grants ending later this year, NHGRI sought to consider the new opportunities for genome sequencing. In traditional NHGRI fashion, we convened a program review meeting in July 2014 that helped to detail several key applications for large-scale genome sequencing going forward, including: (1) determining the genomic contributions to rare and common disease; (2) improving our understanding of how genomic variation influences genome functions; (3) tackling new questions in comparative and evolutionary genomics; (4) assessing the utility of genome sequencing in clinical practice; and (5) understanding genotype-phenotype relationships.
Here, I highlight the first two Requests for Applications (RFAs) resulting from this strategic planning effort. The first RFA is for the establishment of the Centers for Common Disease Genomics (CCDG), which aim to identify genomic variants that contribute to common human diseases, explore a range of common diseases with different underlying genomic architectures, and develop resources for multiple common disease research communities. The second RFA is for a renewal of the Centers for Mendelian Genomics (CMG), which aim to discover the genomic variants underlying several hundred human disorders, called Mendelian disorders, and to develop and share methods to enhance efficiency for others to discover such variants. Such knowledge would facilitate rapid and accurate diagnosis of these disorders and might lead to new therapeutic approaches.

These first two RFAs illustrate how our GSP has naturally evolved over the years to now strongly emphasize the use of genome sequencing for studying human diseases. Consistent with the conclusions of the July 2014 meeting, NHGRI plans to fund other areas involving genome sequencing- watch for a discussion of these potential new initiatives during the Open Session of the National Advisory Council for Human Genome Research meeting at 10:00 AM (Eastern Standard Time) on February 9, 2015 at genome.gov/GenomeTVLive/.
The Centers for Common Disease Genomics (CCDG) and the Centers for Mendelian Genomics (CMG) RFAs are available at grants.nih.gov/grants/guide/rfa-files/RFA-HG-15-001.html and grants.nih.gov/grants/guide/rfa-files/RFA-HG-15-002.htm, respectively. Applications for both RFAs are due April 7, 2015.
I find it striking to compare what the GSP will do over the next 4-5 years compared to what it was doing a decade ago. The fundamental differences in these two time periods truly reflect the monumental progress that the field of genomics has made in capitalizing on its advances in technology and knowledge.
 |
Posted: February 4, 2015
