Nanopore DNA Sequencing
Definition
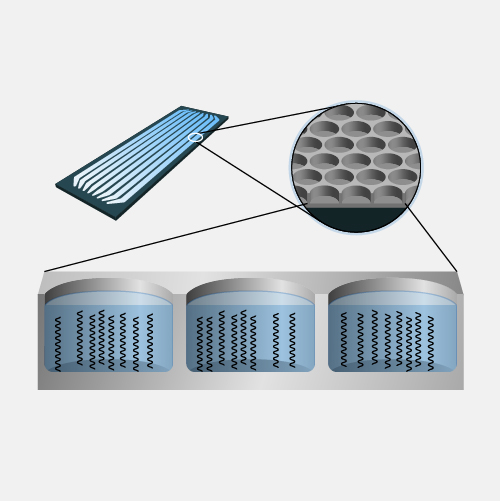
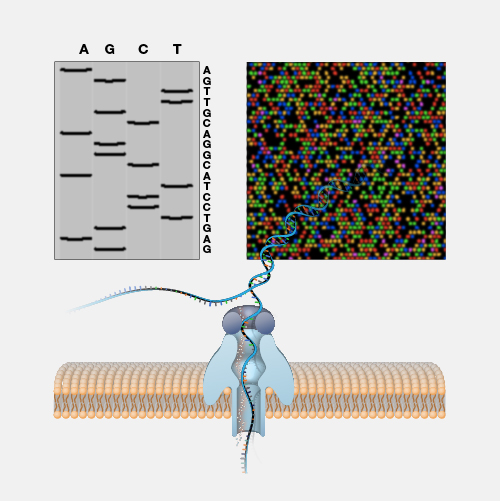
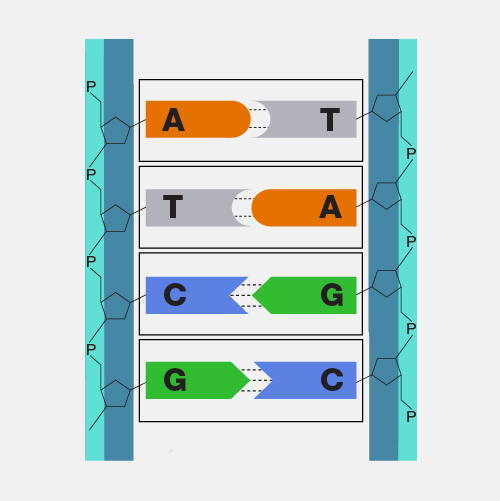
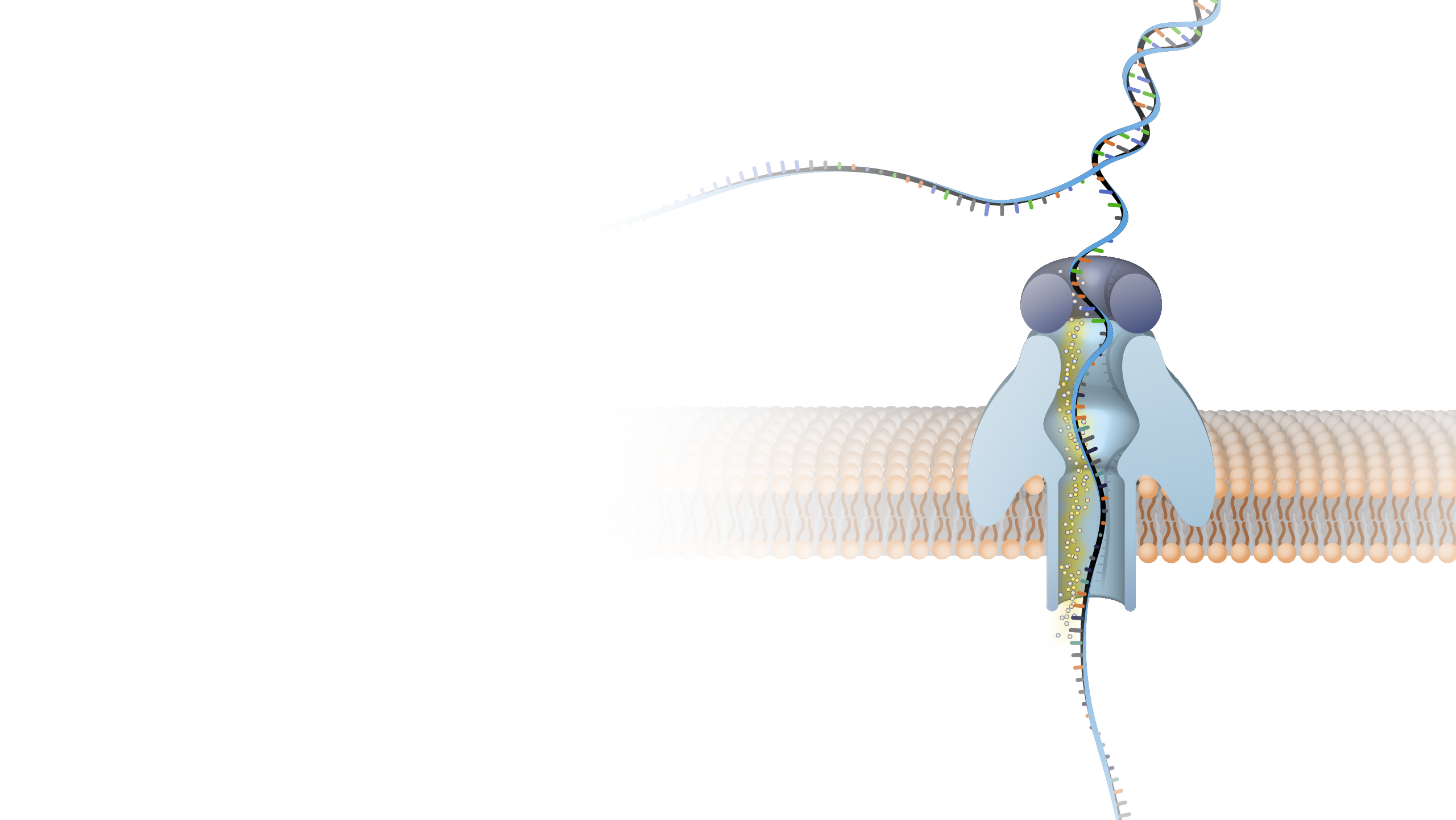
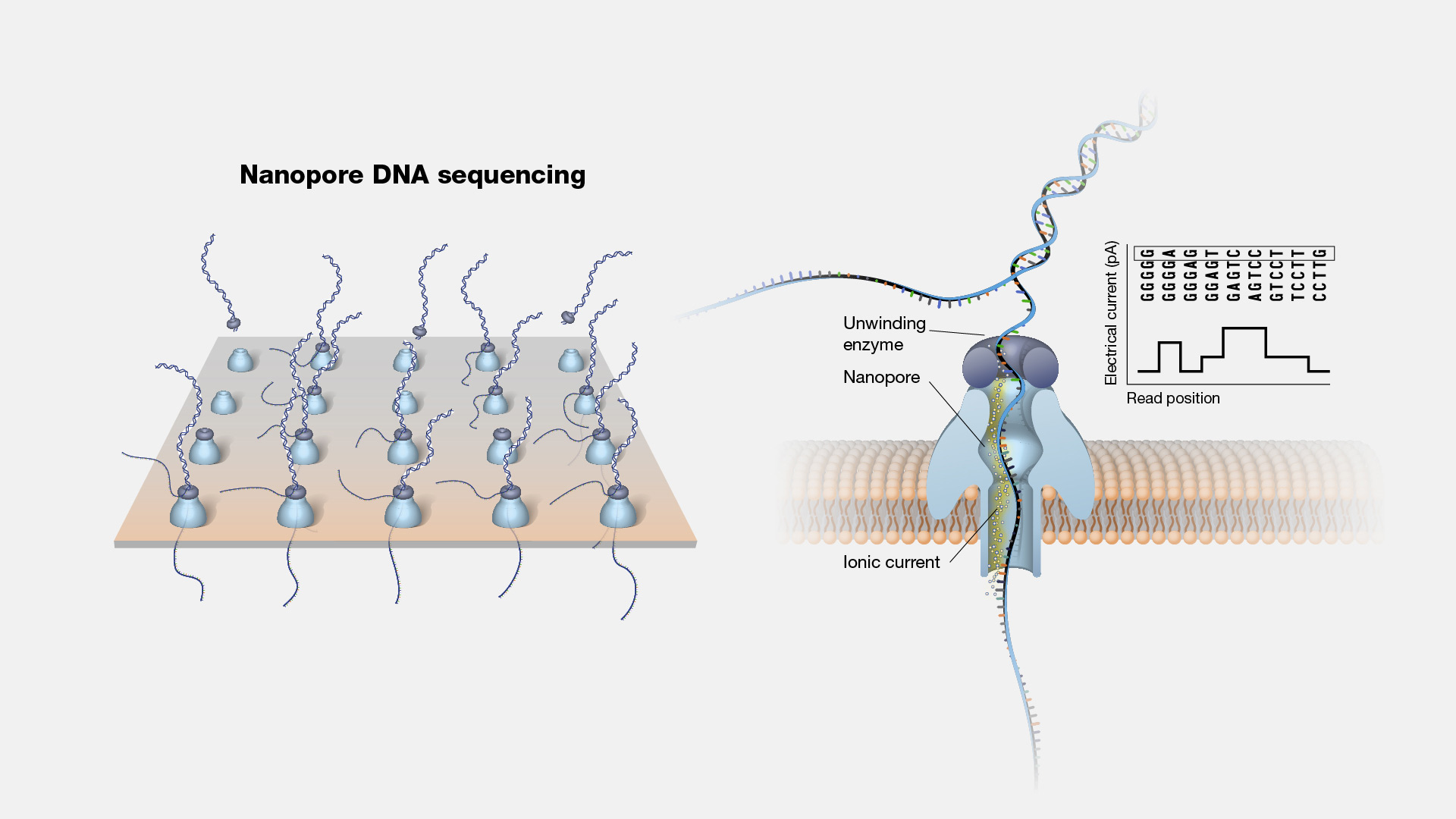
Nanopore DNA sequencing is a laboratory technique for determining the exact sequence of nucleotides, or bases, in a DNA molecule. The sequence of the bases (often referred to by the first letters of their chemical names: A, T, C and G) encodes the biological information that cells use to develop and operate. Nanopore DNA sequencing involves reading the code of single DNA strands as they are threaded through extremely tiny pores (nanopores) embedded within a membrane. As the DNA moves through the pore, it creates signals that can be converted to read each base. This approach offers a low-cost, rapid process for studying long stretches of DNA.
Narration
Nanopore DNA Sequencing. Early techniques for sequencing DNA relied on breaking the genome into many tiny pieces, creating vast copies of those pieces, sequencing them, and then putting the resulting data back together with computers. Nanopore DNA sequencing, however, threads one DNA strand through a really tiny nanopore and then detects minuscule changes in electric current to determine the different nucleotides of DNA. A major advantage of nanopore DNA sequencing is the ability to produce ultra-long reads — much, much longer than that produced by earlier techniques. It also detects the nucleotides from a single DNA molecule rather than many. These advantages have helped scientists work out complex regions of the human genome.