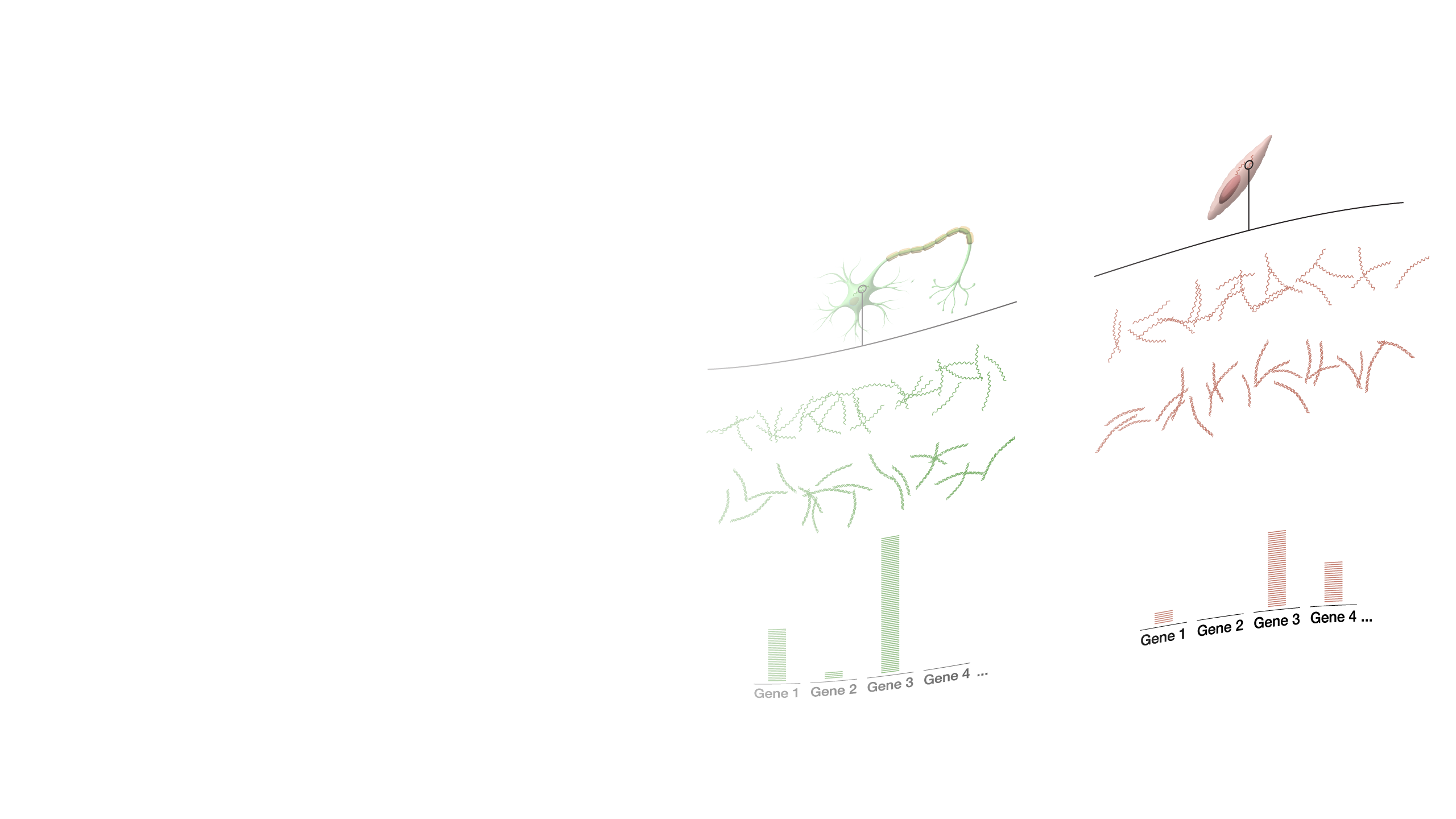
RNA-seq (RNA Sequencing)
Definition
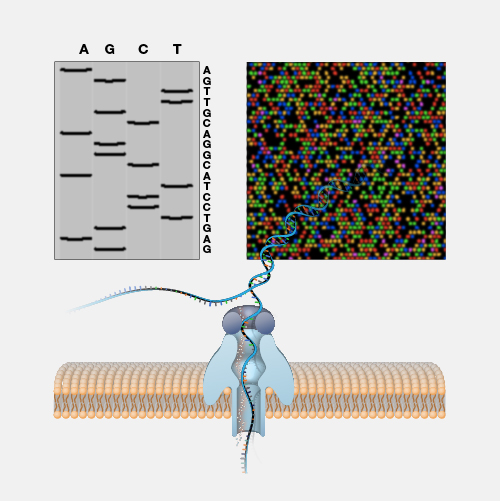
RNA-seq, short for RNA sequencing, is a method for sequencing an entire set of RNA molecules. This method involves isolating RNA molecules in the tissue or cell sample, copying those RNAs into DNA and then sequencing the resulting DNA molecules. RNA-seq helps researchers determine which segments of DNA have been transcribed into RNA and how much a gene is expressed.
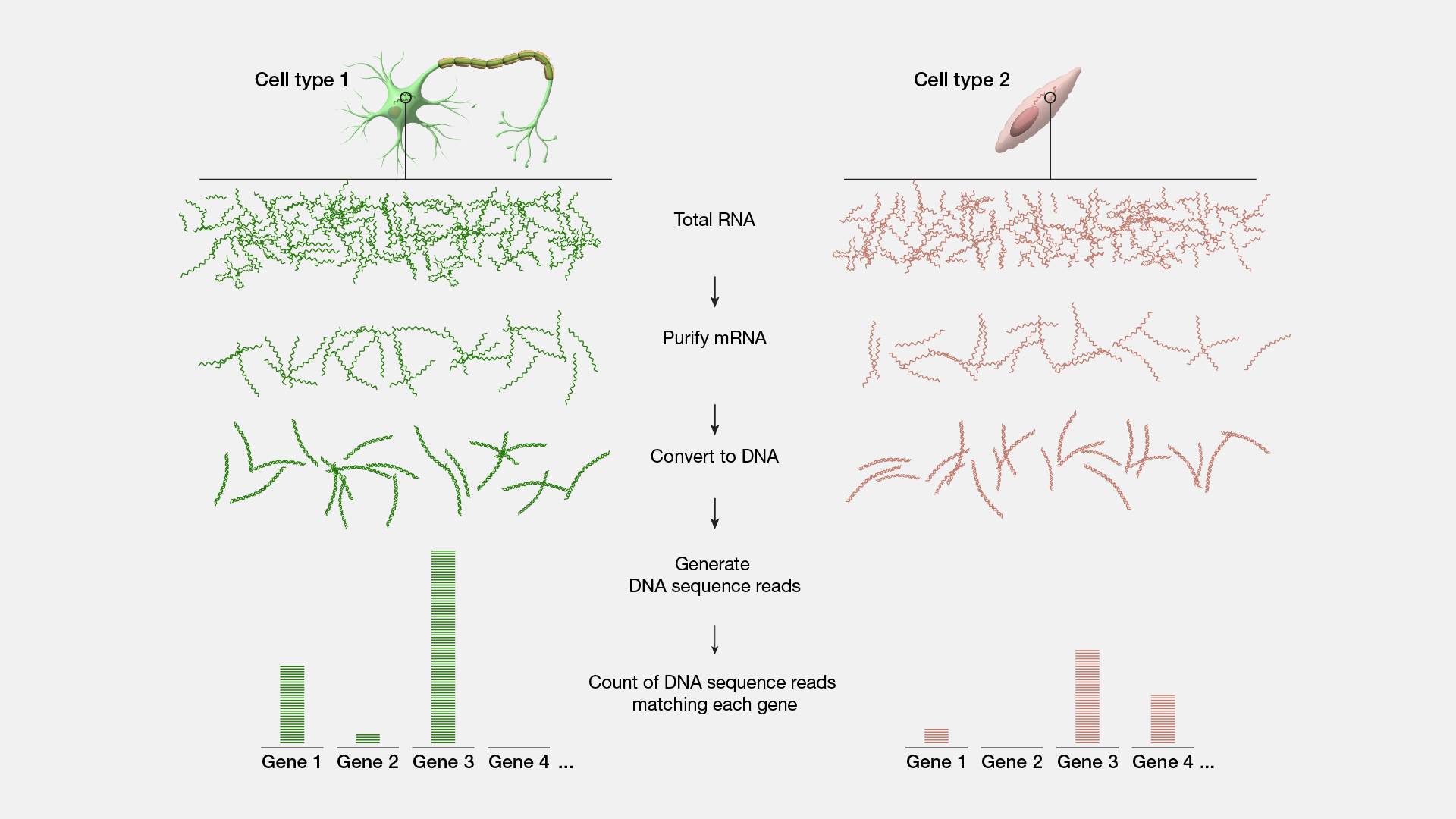
Narration
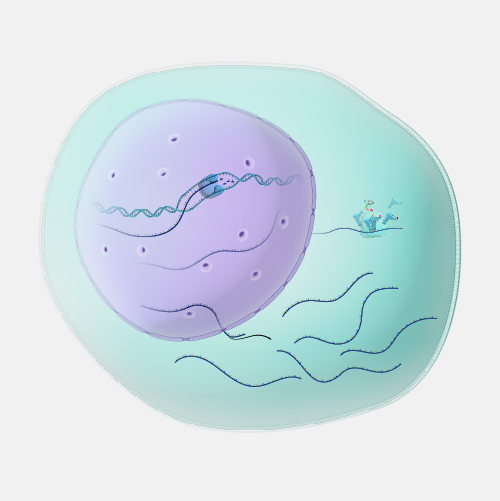
While two different cells within the human body have a similar or identical set of DNA molecules (called the “genome”), the set of RNA molecules (called the “transcriptome”) varies significantly between cells. For example, a skin cell and a brain cell each contain a very different set of RNA. And in addition, again, unlike DNA, the RNA within a given cell is dynamic, rapidly changing over time based on many factors - from time of day to what you ate for lunch; or in more extreme cases, whether or not that cell is healthy or diseased. If we want to measure the transcriptome, the most commonly used method is RNA sequencing often referred to as RNA-seq. RNA-seq is an adaptation to DNA sequencing, which relies on a particular enzyme (called reverse transcriptase or “RT”). RNA-seq starts by isolating all of the RNA from a cell. Then the RT enzyme gets to work and does its job copying all of the RNA into a set of complementary DNA (called “cDNA”). The cDNA is then sequenced using a DNA sequencing machine. The result is data telling you the specific sequence and number of different RNA molecules present in a sample. RNA-seq data is used for all sorts of different experiments and research. In order to better understand the function of different genes, to understand and map the location of different cells within the body, or to measure the environment within a tumor or unhealthy tissues, RNA-seq is an integral and important method within the genomics toolkit.